Fish, genes and chips: freedom of expression from pollution

(Photo: ZFIN and Oregon Zebrafish Laboratory)
As pollution increases, there is a growing need to know what damage these pollutants cause in organisms, how they condition the behavior of the organism and how these effects can be predicted. For the study of these damages, although the studies have focused on several levels, those carried out at the molecular level have materialized, promoted and made essential by knowing the whole genomic sequence of eukaryotic beings (fruit fly, earthworm or human being).
The reason for this importance lies in the function and characteristics of the genome itself, which can be considered as a library that houses all the information (phenotype) to form each organism. This library is organized into books or genes that store information to produce proteins that perform most cell functions. Thus, changes in genes and/or expression of these genes can cause alterations in the normal functioning of the cell.
Regulation of gene expression
Phenotypic differences between species (body architecture, metabolism, behavior...) must be understood on a double level. On the one hand, each species has its own library (genome); some species are endowed with numerous books (genes) and others less. On the other hand, and as we have just known, the genomes of man and monkey are similarly endowed, so another thing must condition the notable differences between these two species. This second variable involves the regulation of gene expression.
To a large extent, the books we have in the library are read in one way or another. That is clear if we look at our colco. Human beings are pluricellular beings and, although all our cells have the same genome, it is difficult to mark the similarities between a hepatocyte and a neuron, since the cells during development, according to the conditions of the medium, express (read) certain genes (books). Therefore, to win in the competition between species we use the metabolic pathways of every moment, always with the aim of obtaining the maximum energy yield.
For example, the inhabitants of the first world do three meals a day. Therefore, to meet our energy obligations, on the one hand we resort to carbohydrate metabolism through glycolysis enzymes, but on the other hand we accumulate lipids through the production of triglycerides. Therefore, we do not need the catabolism of lipids until we feel hungry. When a hunger strike begins, for example, enzyme-producing genes from metabolic pathways involved in triglyceride catabolism begin to express themselves quickly. It is what is known as regulation of gene expression.


To carry out this regulation, multicellular beings are endowed with special proteins, transcription factors, which work in the continuous exploration of the cellular environment. The function of these proteins is to cope with changes in the environment (changes in food concentration, physiological factors, hormones, parasites, diseases...) by changes in gene expression.
All pollutant and toxic compounds present in the environment are therefore an environmental variable for these transcription factors. We will give an example: most animals can distinguish male and female specimens. The main differences between males and females depend on two hormones in most species: testosterone in males and estradiol in females. Estradiol launches representative development alternatives in females using a transcription factor called estrogen receptor. This causes the production of vitelogenin, the most important reserve lipoprotein of the oocytes of female fish (also in other groups). They produce female fish, not males, although they also have the gene that produces vitelogenin.
In large quantities, compounds with a chemical structure similar to estradiol arrive in our rivers, such as alkylphenols of soaps used in our cleanings, phthalates used in plastics or artificial hormones we take in birth control pills.
These compounds transcribe through the estrogen receptor the gene that encodes vitelogenin in males. But the vitelogenin gene is not the only one that responds; estrogenic compounds given to a male influence the expression of hundreds of genes. As a result, they exist in the river world, where male fish produce female-marking proteins, in which, along with sperm, oocytes have also been found in some cases. This example therefore highlights the significant effects that some compounds of daily use may have on the populations of many organisms.
Toxicology and genomics together
One of the current objectives would therefore be to know the obligations of all these genes of different genomes in the cell. Although we are still quite far away, year after year we know the genome of more organisms and we know more and more clearly what are the mechanisms of regulation of gene expression. Therefore, toxicogenomics aims to investigate which mechanisms of regulation of gene expression put in place certain pollutants.

It is known that not all individuals exposed to environmental pollutants respond in the same way. This response is limited by internal factors of the host (state of development, genetic, hormonal, metabolic character...) and environmental (concentration of pollutant and exposure times). Therefore, depending on the combination of contaminant, presence of cofactors and genetic sensitivity, two individuals or two cells of the same individual can respond differently to a particular effect.
The response of organisms to a certain toxic substance is a consequence of the interactions of expression of different genes. Therefore, in recent years more and more laboratories are studying these genes. Hence toxicogenomics was born.
In toxicogenomics, the analysis of gene expression, in addition to reporting on the state of the cells, shows us how these cells respond to different stimuli. Therefore, first of all, we should know the genomic sequence of the organism of interest or, at least, the burdens in which the medium variables can be expressed in excess or in excess and in which metabolic pathways they intervene.
In recent years there has been tremendous progress in this field and it is largely known for the different types of compounds, such as those that cause inflammation, estrogenic compounds, immunosuppressants or carcinogens, which modify gene expression. So, for example, when we talk about estrogenicity, we know that the main genes to be analyzed are those that encode vitelogenin (in fish) and proteins of the extracellular matrix of the egg; when we talk about immunosuppressants, interleucine, complement protein or various types of cytokines; and when we talk about carcinogenic, along with the geniuses Bcl-2, p53, c-myc, etc.
In determining the genes to be studied, the work of future toxicological agents would be to investigate the expressive changes of these genes against contaminants. However, in unknown cases, toxicologists should look for new genes that respond to the effects of the contaminant.
Fish use in environmental toxicogenomics

To say that fish are the best organisms when analyzing environmental toxicology is not to say much, since these vertebrates have the body immersed in the water and, therefore, are directly related to the pollutants present in the environment, either through the gills or the hepatic intestinal system. The fish group is a broad group, both in the number of species and in the number of individuals within the species, which makes it a group of organisms with different physiologies, life forms and various adaptation systems.
This diversity is also reflected in the genome, as fish have a much more plastic and changing genome than other vertebrates. Therefore, due to changes in the environment, pollution, especially fish, is more sensitive than many other organisms. Likewise, the use of fish as a model will lead us to better understand the mammalian genome when interpreting the genomes of other taxa. In addition, it should not be forgotten that many species of fish, in addition to their ecological or scientific importance, also have an economic importance, especially in those countries where fish is a regular food.
Thus, we currently work with numerous species of fish, including zebrafish, medaca, carp, trout, salmon and saucer. Although our knowledge of each other's genome is radically different, today (November 2005) we know more than 10,000 gene sequences that are expressed in 14 species of telerosteal fish. In addition, the complete genome sequence of teleosteos Fgu rubripes, Danio rerio and Tetraodon nigroviridis is already known.
Microchips: tools to investigate gene expression
We have fish and genes that we want to study, what to do with them? What is the methodology to follow? DNA microbildumes or microchips are current and future work tools based on the increasingly known holistic analysis of gene expression. These chips consist of hundreds or thousands of DNA probes corresponding to genes of interest attached to a solid support.
The microchips have only the size of the glass doors that are used in the microscope and allow us to simultaneously detect all the sequences that interest us and are known. They can detect additional DNA and RNA molecules marked with different markers. To do this, RNA is extracted from each of the fish organs (such as the liver) treated with pollutants and untreated, and the auxiliary DNA (cDNA) is synthesized by reverse transcription. This cDNA constitutes the cell transcriptome, that is, the collection of the gene expression of that animal's genome at that time. This cDNA can be marked with fluorescent markers, one in green and one in red, in short, in order to differentiate the genetic material of both organisms. The same amounts of cDNA from the organisms we want to analyze and compare are then combined and the mixture is hybridized with microchip. Thus, if at the time of extraction in the treated fish was expressing a lot of vitelogenin and at the time of extraction in which it was not treated, this difference could be detected by the different colors that the chip will present for that gene.
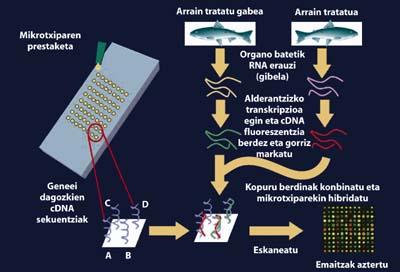
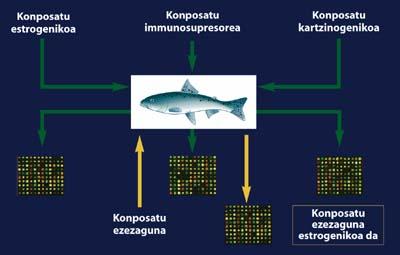
In laboratory experiments that have been conducted throughout the world, certain types of fish have remained exposed to certain toxic compounds. In these cases microchips have been developed on one of the fish organs, so it is already known the gene that responds to the exposure of these species to this type of pollutants.
These expression patterns do not differ much from those known in mammals. At present, if any of these organisms that we know how the gene expression pattern changes are under the influence of an unknown compound in the environment, we would have the ability to identify the type of contaminant with respect to the expression patterns we know. Thus, before this pollutant can cause harm to living things, we would have the opportunity to take local measures to prevent them. In the same way, once new drugs and compounds are manufactured, we can foresee the changes that these can produce to the organisms before they are put on the market.
Molecular mechanisms of pollutants and environmental quality analysis
In recent years, laboratories that apply chips to fish are increasingly numerous, both in laboratory experiments and in rural populations. Studies in the UK, for example, are trying to determine new molecular biomarkers that respond to specific contaminants and define their mechanisms.
Thus, the gene expressions of the Platija fish that inhabit the arid ria called Alde and the contaminated Tyne Estuary have been compared. To do this, after cloning in the dish genes related to different types of stress in other species, they have designed a microchip of 13,000 sequences. After hybridizing the cDNA of fish from both areas with chipis, significant expressive differences have been found in one hundred sequences. The most representative of those induced in Tyn have been related to the presence of high concentrations of aromatic polycyclic hydrocarbons in the medium CYP1A1, paraoxonase (phase I enzymes of detoxification metabolism), UDPGT and aldehyde dehydrogenase (phase II enzymes). The changes found in the other genes, however, respond to global stress.
In addition, they have observed that estradiol induces, among others, the genes that encode vitelogenin and choriogenin. Some of these expressive changes have been described in other recent laboratory experiments. For example, in the cymbal males maintained for 21 days under ethinyl estradiol, more than 200 genes manifested different expression in treated animals. It should be noted that among these genes is induced the expression of the gene encoding vitelogenin, UDPGT and the protein of the extracellular oocyte matrix, ZTP.
DNA microchips are therefore powerful tools to characterize the toxicological properties of chemicals. Like any other growing technique, numerous research is still needed to define and consider the use of DNA microchips in the field/environment from the point of view of ecotoxicology. However, microchips are accurate photographs of the gene expression of organisms and can clearly show how fish physiology is reorganized under the effects of changes in the fish environment.
BIBLIOGRAPHY




