2008/02/01
239. zenbakia

eu es fr en cat gl
Aparecerá un contenido traducido automáticamente. ¿Deseas continuar?
Un contenu traduit automatiquement apparaîtra. Voulez-vous continuer?
An automatically translated content item will be displayed. Do you want to continue?
Apareixerà un contingut traduït automàticament. Vols continuar?
Aparecerá un contido traducido automaticamente. ¿Desexas continuar?
Classification of living beings: from morphology to DNA sequencing
Text created by automatic translator Elia and has not been subsequently revised by translators.
Elia Elhuyar
For the research and understanding of any system, a certain ordering of this system is almost essential: separation, designation and classification of system elements. This is what taxonomy does: name and classify living beings. But the way to make this classification is changing.
Classification of living beings: from morphology to DNA sequencing
01/02/2008 | Etxebeste Aduriz, Egoitz | Elhuyar Zientzia Komunikazioa

(Photo: N. N. G.W. from Kurz Rouse/Tree of Life Web Project)
a.C. Around the year 350 Aristotle made a classification of living beings. They were divided into two groups: the animal kingdom and that of the plants. He also used the term species. However, the real taxonomy XVIII. It can be said that he was born in the twentieth century by the Swedish Lineo. Lino wanted to classify plants, animals and minerals created by God.
According to the mentalities of his time, God created things with a plan or order. And it seems that the goal of Line was to find that order. Thus, he studied morphological characteristics and created a hierarchical classification, taking into account similarities and differences. In his book Species Plantarum he described plant species, grouping similar species in genera - with common characteristics - and, in turn, genera in families. In addition, to designate each species, he used the Latin binomial name --Genus specia-. At present, these types of names are still used by scientists from all over the world to call a species in the same way, and the bases of the current taxonomy are also those established by Line.
However, with the theory of Darwin's evolution, the philosophy of the classification of beings changed. In fact, according to the theory of evolution, from some species to others emerged throughout the history of life, so that, more than knowing God's plans, since Darwin has sought a classification that reflects this history of life -- filogenia---.
The classification of living beings adopts the form of a tree called a phylogenetic tree. At one end of the tree are the ancestors or ancestors of all living beings, and at the other end all current species. In this tree, the species are not collected in gender only by their similar characteristics, but by their origin, and therefore have similar characteristics.
In the footprint of morphology

These four animals are mollusks. This means that the morphological characteristics of the four have arisen from the evolution of the same ancestor.
macrophile; sarae; D. Burdick/NOAA; J. Petersen Petersen
Morphological characteristics have long been the principal traces of taxonomies in order to build the tree of life. But the classification of beings by these characteristics is not a simple task. For example, we will see at first glance that snails and slugs have similar characteristics, or chipirones, txokos and octopus. But to see that all of them, on another level, have the same origin, one must look more thoroughly. They are all mollusks because they have the same basic organizational model. That is, even if it seems a lie, the characteristics of snails, bazos, chipirones, octopus or mussels can be explained by the evolution of the supposed characteristics of a hypothetical ancestor of all molluscs.
To reach this type of conclusions, we must analyze something more than the characteristics that are in sight. For example, many times useful clues are found in the early stages of the development of living beings. For example, our ancestors had a tail and we also have an embryonic state. Or the embryos of the whales, like the rest of mammals, first develop four legs and then almost disappear.
In addition, you have to be very careful with fake fingerprints. In many cases, throughout evolution, various species have obtained similar characteristics by different pathways. This is what is known as evolutionary convergence, such as bird wings and bats, or the fine-shaped structures that aquatic animals have developed differently. Sometimes it is clear that beings of similar characteristics have no relation, since they have many other different characteristics. However, on other occasions, it is not easy to know if some characteristics have or do not have the same origin, and if they are used to build phylogeny, it can be thought that groups away from each other are close.
On the contrary, morphologically simple beings, like nematodes, offer very few traces. This type of groups cause large headaches to the taxonomics. And let's not say in the case of microorganisms. Many times it is very difficult to classify them by morphological characteristics.
Thus, although much has been worked on the construction of phylogeny based on morphology, it has its limitations. And in some points there are great difficulties to advance only with morphology.

Winged animals have developed their wings by different ways.
Archive; Dr. Hemmert; Archive
History of life in the genome
In the last 30 years, the molecular techniques based on DNA have been revolutionary in the classification of living beings. In short, the changes in evolution are reflected in the genome of living beings, so the information contained in the genome can be of great utility for the study of phylogeny.
Phylogenomics is the study of phylogeny from large amounts of genetic data. Automatic sequencing techniques of PCR and DNA, for example, allow to amplify and sequence many genes in a relatively simple and fast way. In this way, genetics are hitting and sequencing DNA. They have already sequenced complete genomes of about a thousand species and each month sequenced genomes of more species are being published. And by comparing the sequences of these genomes - or of some genomes - molecular phylogenies are constructed.
Over time, mutations or changes in the genome occur. These changes, sometimes, will have decisive consequences and will progress or not depending on the natural selection. But many other changes have no significant consequences. For example, in genes there are regions that do not encode proteins: intronas. The changes that occur in these zones, in most cases, do not affect the selection, which is why they accumulate throughout the evolution. The same occurs with mitochondrial DNA, where many changes accumulate, mitochondrial DNA is widely used in molecular taxonomy. Groups close to each other will experience similar cumulative changes, so their DNA sequences will be very similar.
Molecular techniques have allowed great advances in the classifications of microorganisms. The study of the diversity of bacteria performed 10-15 years ago through DNA sequences gave surprising results. The sampling of different media and the amplification and sequencing of the ribosomal RNA genes (SSU) allowed us to observe that the number of varied fire sequences was between 20 and 100 times higher than what was observed until then in studies based on cultures. In addition, although some of these sequences were similar to those of the known bacteria of cultured cultures, others were quite different to suggest different evolutionary lines without known representatives.

Molecular techniques have gained great importance in the taxonomy of microorganisms.
ANDÉN
And, despite the controversies it generates, in the rest of the living beings, taxonomy based on DNA is becoming increasingly important. It has its problems, but also its advantages against classical taxonomy. On the one hand, they are very useful to clarify many things that cannot be clarified with morphological characteristics, and on the other, much faster than performing morphological studies.
In spite of the debate, it seems that in taxonomy the genetics will be advanced to the morphology and, henceforth, in the sequences of DNA we will be able to read many passages on the history of life.
Raúl Bonal: "the debates arise because classical taxonomists have seen molecular phylogeny as a threat"
In the Department of Entomology of the Natural History Museum in London, a team of researchers led by Alfried Vogler investigates the phylogeny of beetles based on DNA sequencing. In fact, they have recently published a phylogenetic tree made with 1,900 species of beetle. Dr. Raúl Bonal is a researcher of this team who has responded very willingly to our questions.
It seems that molecular techniques are strengthening taxonomy. That's undeniable, isn't it?
Yes, it is true, molecular techniques are revolutionizing taxonomy. Thanks to the techniques developed in recent years, it is currently relatively easy to sequence DNA. And by comparing the changes that have occurred throughout evolution in different genes, beings can be classified.
Molecular techniques allow to solve to a large extent some problems of classical taxonomy, such as evolutionary convergence. However, molecular filogenias also present similar problems: sometimes a change produced by a mutation can be restored by another, and in these cases we will not detect any change, where in reality there have been two changes.
On the other hand, it is possible to know the biodiversity of a place quickly, sequencing and classifying the DNA of the beings found. In addition, it is very useful to identify some of the phases of the life cycle of some beings. For example, larvae of many insects lack identification key and can be identified by comparing their DNA with the DNA of identified adults. The same with different species (cryptic) that cannot be separated morphologically, or that, because they are morphologically different, are the same species. In these cases, DNA sequencing is very useful.

(Photo: S. S. Huts)
But molecular taxonomy generates great controversies...
I believe that controversies are due, rather than technical problems, to which classical taxonomists have seen molecular phylogeny as a threat. For example, a classic taxonomist, expert in a group of insects, whose work must be respected and esteemed, does not like scientists from another field, such as genetics, to start talking about new species. They feel they are invading their own.
In addition, the appearance of new species is at high speed and often used in filogenias without giving name or describing them. That doesn't like the classics anything. However, the number of new species concentrated in some parts of the planet is so high that it is very difficult to make a morphological description prior to the construction of phylogeny.
In short, I think it is a confrontation between different schools and professionals.
One of the main problems of taxonomy has always been the problem of the correct definition of the species. Will genetics solve this problem?
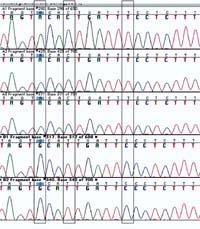
In this DNA chromatography, the DNA sequences of six individuals can be compared.
(Photo: Molecular Systematics Group, Dept. Entomology, NHM)
Defining what a species is is very difficult. The classical definition 'hard' says that the different species cannot be crossed and that if the following hybrids are crossed they must be sterile. But in reality this is not always fulfilled. One example is the famous Darwin's txontas, which, however, are considered different species. Therefore, the species is defined from a group of populations that share characteristics and interact spatio-temporarily.
Molecular phylogenies take into account this last definition. In the phylogenetic tree, made by comparing DNA sequences, it is possible to calculate, by statistical calculations and models of molecular evolution, where the speciation begins. That is, you can draw a line that cuts the branches of the tree to a certain height, and the individuals that remain there together belong to the same species. On the contrary, the branches that are formed from there indicate the genetic diversity of the same species.
The concept of species is debatable in biology, but the effectiveness of this form of species determination through DNA sequences is demonstrated. In the cases in which the test with known species has been conducted, results have been obtained very similar to those obtained in a classical manner.
Will the classifications based on DNA leave aside morphology?
Maybe because I think they have to get to talk to us or I think they don't. It is true that genetics is much faster. Many people do not realize how many species are to be discovered. In insects, for example, it is estimated that ten million species can exist and no tenth is known. In this sense, morphology cannot compete with genetics.

The beetles of the genus Curculia pierce and lay their eggs in the acorns.
(Photo: R. R. Bonal)
But I do not think that we must abandon morphology. The morphological description of the species is important. Filogenias can also be performed, combining molecular and morphological characteristics. In addition, although the sequencing of DNA is increasingly available to scientists, it is not available to amateur naturalists, so it is good to have morphological keys. In addition, the knowledge that classical taxonomists have cannot be lost. And here I want to denounce that is being lost because in many museums there are no new places for this type of professionals.
What do you think about the genetic barcode?
It is an interesting and attractive idea, almost as it is done in supermarkets: to pass a being by a detector and say what species is the detector...
As raised at the beginning, it has several problems. The idea was to sequence the same gene for all species and thus use a sequence determined as identifiers of each species. But species are defined according to the classical classification. In this way the problems of morphological taxonomy are maintained and the same mistakes are made. In addition, for cryptic species that do not differentiate morphologically but have a different genetic sequence, what criterion should be taken?
I think it is more appropriate to make phylogeny based on DNA, and thus use the classification made to fix the barcode corresponding to each species.

DNA studies are very useful for identifying larvae of many insects.
(Photo: Entomart)
You work with coleoptera. It is the largest group of insects with a great morphological diversity. What advantages do molecular techniques offer in the case of coleoptera?
For the characteristics mentioned, the group of coleoptera is one of the groups that stands out most for the advantages mentioned above. With such a large number of species, many are not known or poorly known, and in classical taxonomy there are many lagoons. In the tropical regions this ignorance is very evident. In addition, for many larvae there are no identification keys (in some cases the larvae have never been collected). All these problems can be solved thanks to molecular taxonomy.
What exactly do you investigate?
In our group there are people who work with different groups of coleoptera: beetles more coprofos, granívoros, aquatic... We build phylogenetic trees from the DNA, we determine species... And, in addition to investigating the tree, evolutionary patterns of genes are also investigated.
On the other hand, from phylogeny, we also investigate the evolution of certain morphological, ecological, reproductive strategies, etc. I myself am investigating the evolution of the body size of the genus Curculio. The larvae of these insects are parasites of acorns. The first step is to build a molecular phylogeny and the next step, to analyze the evolution model that has followed the body size according to this phylogeny.
Precisely, through these studies I have been able to verify some of the advantages of molecular taxonomy. In Europe there are five species of the genus Curculio, well known. In the United States there are more species, but they are well studied. In Central America the question is very different. In a sampling conducted in Mexico for a month, 20 species were found, according to DNA studies, and we estimate that taking into account the missing areas for sampling, they can be hundreds. If we had to do this diversity study from morphology, we would need a long time. In addition, through DNA we can introduce larvae into phylogeny which are much easier to collect.
Etxebeste Aduriz, Egoitz
Services Services Services
239 239
2008 2008 2008 2008 2008
Services Services Services
036 036 036
Biology; Evolution; Genetics
Article Article Article
Others













