They create a new genetic editing technique, more specific than CRISPR.
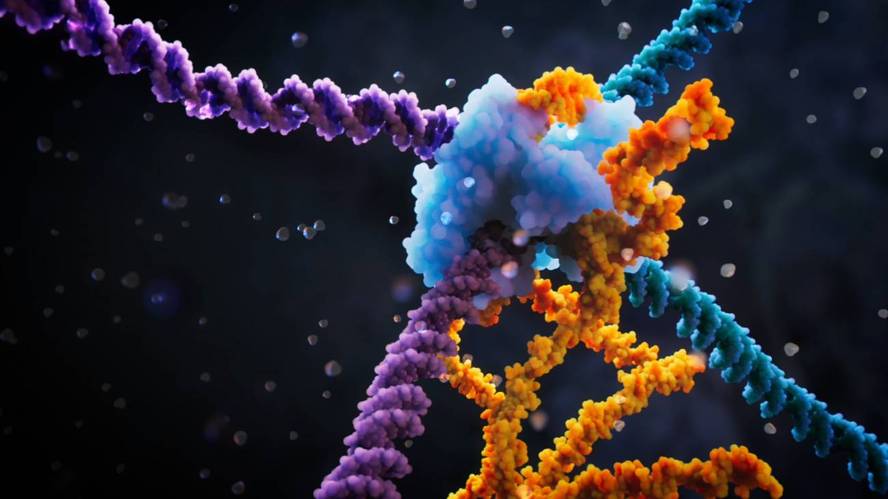
A technique called RNA-Bridges allows inserting, inverting or removing long DNA sequences in specific positions of the bacterial genome. It is more effective and accurate than current methods, but it has not yet been shown to be useful in human cells.
The technique, developed by a team of researchers from the United States and Japan, has been developed in Nature through two articles. The first describes the RNA bridges: It is a RNA conductor capable of identifying and associating the exact position that one wants to change in the genome and the donor.
In the second article, cryochroscopy has been used to unravel the molecular structure of the complexes that emerge in the edition of RNA-bridges. The IS110 enzyme participates in the complex. Thus, they have criomicroscopically discovered the structure of all the elements of the complex that intervene in the process at every step, that is, how they know the IS110 enzyme and the RNA bridge the DNA target and DNA, how they cut the two DNA chains, how they cut and glue those parts and how they do the same with the rest.
RNA bridges are found in bacteria and archos, so researchers have inspired them to create the RNA bridge method. If it is useful in mammals, it offers many possibilities, as it is simpler and more precise than CRISPR. However, they have warned that the CRISPR method is highly developed, so a great deal of work will have to be done to reach and demonstrate that it exceeds the technique of RNA bridges.