Hidden uses of DNA
DNA, in addition to being a foundation of life, has become an icon of current science. However, many do not know that DNA has begun to transcend the biological context due to its exceptional properties as nanomaterials. This article analyzes the capabilities of DNA and dna-based nanotechnology applications.
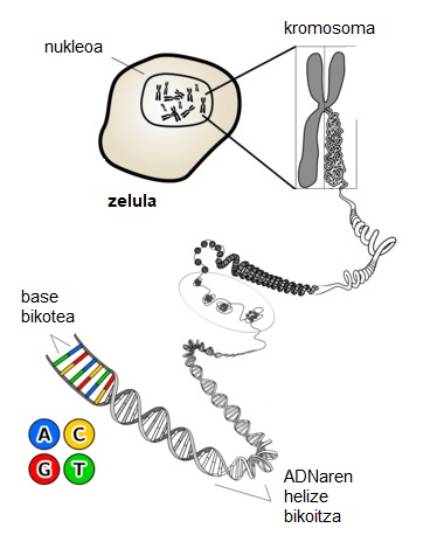
Deoxyribonucleic acid, what a mouth knot! In addition to the mouth, the structure it adopts in our cells is complex, since in a few cubic micrometers a DNA is introduced that can reach a length of two meters. However, the image we can imagine is far from the order that DNA presents, as it fits perfectly into the nucleus of the cell with complex four-letter instructions that form DNA (Figure 1).
According to experts, the essential molecule for life emerged in nature billions of years ago, and until the beginning of the last century its four units of repetition were not identified: adenine (A), guanine (G), cytosine (C) and thymine (T). Later, XX. mid-century described the most common structure of DNA: the double helix. Since then, nucleic acid research has taken the speed of light and developed until the end of the century the field of research for DNA nanotechnology has been developed. DNA would no longer be an essential part of living beings, but would become a material of many other non-biological applications[1].
DNA Faculties
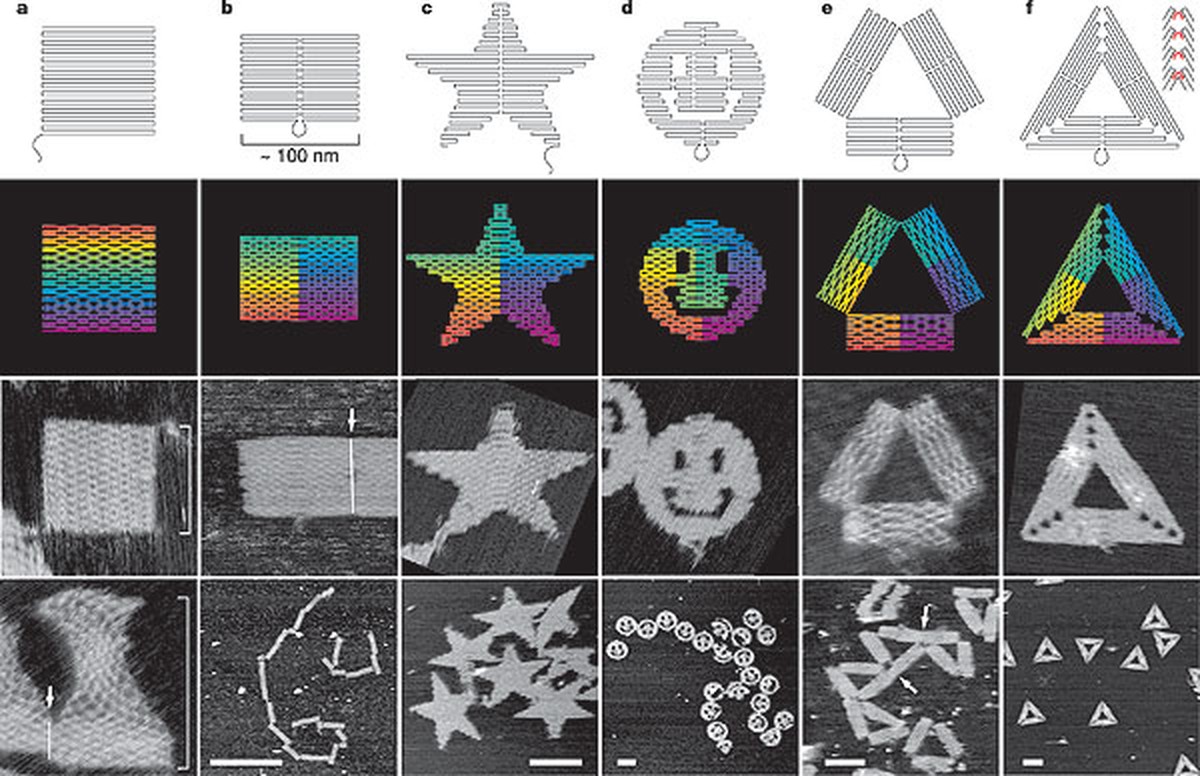
The first proposal was the application of DNA outside nature to form nanostructures. In fact, DNA generates links between its chains very specifically, using hydrogen bridges: Combining letters A with letters T and letters G with letters C. This makes it possible to design precisely at nanoscale, as is the case with the DNA origami technique[2]. This technique, presented a decade ago, and taking into account a simple computer program and DNA connections, you can create the desired constructions, as can be seen in figure 2. However, DNA is not a simple brick to create nanostructures, its letters include other functions.
Imagine that you want to build a house with the same type of material, but for the groups of bricks that you want to do a specific task like catching pathogens from the air. This can be done with dna-generated nanostructures. In fact, DNA has the ability to detect molecules or macromolecules specifically and we can design the desired compound. How is it possible? As already indicated, DNA has only four types of repeating units: A, G, C and T. But by changing the order and length of the sequence of these letters, we can get unlimited combinations. Through the SELEX procedure[3] we can obtain a sequence that generates links with the target molecule.
The type of DNA with sensing capacity is called tamer, and DNA has an extra ability to accelerate chemical reactions. The latter are called DNA or catalytic DNA enzymes and, as with the tamers, the SELEX procedure provides the necessary sequence for each reaction condition. DNA aptamers and enzymes are part of the functional DNA group, greatly increasing the value of DNA as nanomaterials. However, DNA is not a perfect material, as certain properties necessary in nature can set limits to nanotechnological DNA.
In fact, DNA is a charged molecule, negatively charged by phosphate groups, which needs contraions to maintain a stable conformation. On the other hand, DNA degrades rapidly in the aquatic environment or in the presence of enzymes in the air. Inside the cell, DNA is in a very controlled environment, where water activity and contact with other molecules are limited, although when given some other application, sample loss can occur in a few days or months. On the other hand, in addition to water, DNA is stable and functional in a few solvents, since it is known that water and life have always been in close relationship with our planet.
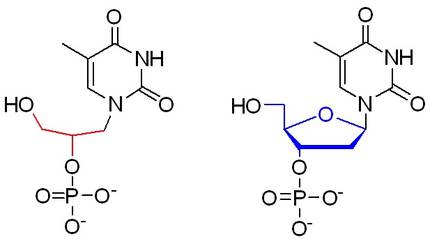
These limitations may be partially overcome, but many studies remain to be developed. There are two areas of research to deal with unforeseen situations by nature: Transformation of the medium in which DNA is dissolved or changes in DNA structure. As for the former, it tries to reduce water activity by simulating the cellular environment[4] or other solvents such as ionic liquids[5]. On the other hand, there are in nature two types of nucleic acids, DNA and ribonucleic acid (RNA). In the last decade, the first synthetic nucleic acids[6] were presented to address the problems of solvent and degradation (Figure 3), although the problem of high economic cost persists and it is easier to make chemical changes in the DNA molecule than to use a new nucleic acid.
Use of DNA outside the biological field
Currently, most applications are medical, including the addition of nanostructures and DNA nanoparticles for the transport and administration of drugs within the body or as biological markers. However, the use of DNA, apart from biology, has much to do with many other areas.
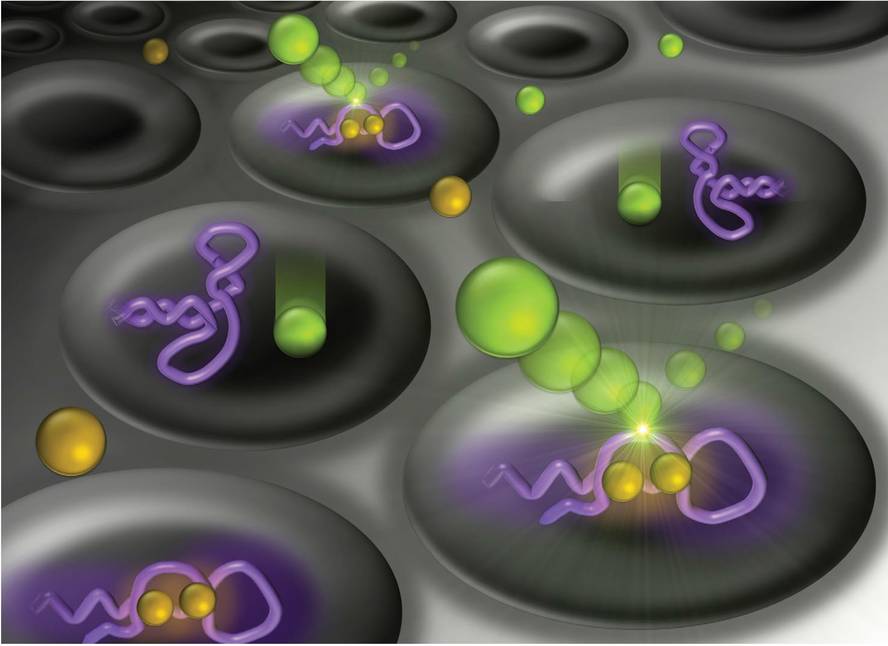
The sum of forces is the cause of extraordinary capacity, and the sum of the structural properties of DNA and perceptive and catalytic capabilities results in applications that cannot be achieved with other materials such as stimulus responses. Membranes are porous materials that separate two media. Since the size of the membranes' pores in specific industrial processes is a few nanometers, it is often very interesting to be able to open and close the pores to your liking. Normally the closing of the pores is done by changes in the whole environment: pH, temperature, pressure, etc. However, DNA designs allow a local agent to control the permeability of pores. More specifically, in the pores nanostructures formed by tamers can be placed to close and open the pore with a specific molecule, since the DNA conformation changes when detecting the molecule[7] (surface image).
DNA enzymes, as already mentioned, accelerate chemical reactions and, in addition, provoke the quirality to achieve the so-called asymmetric catalysis. This type of catalysis is very important and has a high demand in the market, since many chemical compounds are enantiomers. In a couple of enantiomers of one molecule, one would be the mirror image of another and could not overlap (as with our two hands). The point is that one of the images in the mirror usually has very interesting applications and the other does not, so it is much more appropriate to synthesize directly only one of the enantiomers than having to separate them later. Many years ago we know that nature uses proteins to achieve asymmetric catalysis and, in the past decade, the Nobel Prize team Feringa made the following discovery: DNA can also be used for this task thanks to the winding of its double helix[8].
Moving away from the field of chemistry and biochemistry, functional DNA research is declining, but there is great interest in creating nanostructures and using DNA to store information. Our personal information is stored on hard drives with silica chips or magnetic tapes. The amount of data that can be stored in a certain size has increased considerably in recent years, approaching a limit in which physically it will not be possible to increase that compaction. DNA, for its part, is a much denser material in terms of data storage: in a gram could be introduced around two petabytes, that is, in a few grams we would have all the contents available on the Internet. And, in any case, from theory to practice there is great work. Currently, Microsoft and Food companies are the ones that are making the most investments in this application, and although only the first steps have been taken so far, they have already presented their first DNA hard drive[9] (Figure 4).
Thanks to multidisciplinary research, there are still open doors for developing DNA applications, building more robust nanostructures, and leveraging the capabilities of tamers or DNA enzymes. To prevent the knots of nucleic acids from becoming hair in the mouth, we must continue to investigate, since this material necessary for life has much to say outside the cells and it is in our hands to release the knots and profitability options.
Bibliography
[1] Seeman, N. C. & Sleiman, H. F. (2017). DNA nanotechnology. Nature Reviews Materials, 3. DOI: 10.1038/natrevmats.2017.68
[2] Rothemund, P. W. C. (2006). Folding DNA to create nanoscale shapes and patterns. Nature, 440(7082). DOI: 10.1038/nature04586
[3] Wang, T. & Veedu, R. N. (2019). Available technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnology Advances, 37, 28–50. DOI: 10.1016/j.biotechadv.2018.11.001
[4] Nakano, S., Miyoshi, D., & Sugimoto, N. (2014). Molecular Structure Crowding on the Structures, Interactions, and Functions of Nucleic Acids. Chem. Rev., 114, 2733–2758. DOI: 10.1021/cr400113m
[5] Olave, B., & Schäfer, T. (2018). CHAPTER 17 Functional DNA in Ionic Liquids. In Ionic Liquid Devices (pp. 423–444). The Royal Society of Chemistry. DOI: 10.1039/9781788011839-00423
[6] Holliger, P. et. to (2019). A synthetic genetic polymer with an uncharged backbone chemistry based on alkyl phosphonate nucleic acids. Nature Chemistry. DOI: 10.1038 per month
[7] Schafer, T., & Ozalp, V. C. (2015). dna-aptamer gating membranes. Chemical Communications, 51(25), 5471–5474. DOI: 10.1039/C4cc09660f
[8] Fering, B. L. et al. (2010). Catalytic enantioselective syn hydration of enones in water using a dna-based catalyst. Nature Chemistry, 2(11), 991–995. DOI: 10.1038/nchem.819
[9] https://youtu.be/tBvd7OSDGgQ, https://www.youtube.com/watch?v=60Gi5lqL-dA